A Nanoscale Test Tube
Looking at enzymes through the eye of a nanopore
The study of single molecules is a relatively new field of research but covers a wide range of systems from single atoms to complex biological systems. In multiple turnover enzymatic reactions molecules are not synchronized in their dynamic behaviours and, at a given point in time, each enzyme molecule is at a different stage of the reaction sequence. When single molecules are observed, synchronization is not an issue and the observation of transient subpopulations, that may be lost in ensemble measurements, becomes possible. Information about the dynamic coupling between protein motions and catalysis within a single molecule can be obtained. Inactive enzymes can be ignored because they are simply “not seen”.
Our approach
We are developing a new technology to investigate single-enzymatic reactions by using a protein nanopore. Single enzymes are are trapped within the lumen of the pore and the enzymatic activity or the unfolding kinetics measured by the disruption of the ionic current through the pore. The advantage of nanopore investigations is that they allow the study of native enzymes with a temporal resolution ranging from a few μs to hours.
Analytes binding to DHFR
DHFR is trapped inside the nanopore by the electroosmotic flow (EOF). The binding of the substrate NADPH and dihydrofolate to the protein induce transitions from the open to the closed structure of the protein. DHFR remains trapped inside the nanopore for minutes. Nanopore recording allow the longest single-molecule investigations of analyte binding recorded to date. Proteins inside the nanopore remain folded. Further, the binding constants of the proteins inside and outside the nanopore are identical, suggesting that the nanopore is a suitable test tube for single-molecule analysis.
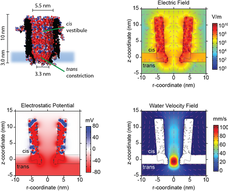
A deeper look: Nanopore properties
We performed simulations to understand the environment of the trapped proteins under an external applied potential.
Inside the ClyA cis vestibule the electrostatic potential at the centre of the nanopore (r-distance = 0) increased almost constantly, after which it rapidly increased near the trans constriction.
Interestingly, the electric field showed two small minima located at ~4 and ~10 nm above the lipid bilayer, where proteins most likely are trapped
ClyA has a strong negative surface charge that under an applied potential induces a directional net transport of water molecules through the pore (the electroosmotic flow, EOF). We found that in the cis vestibule of ClyA-AS the water velocity was relatively constant to then rapidly increase at near the trans constriction.
The EOF is the dominant force that keeps proteins inside the ClyA nanopore
Our Publications
Single-Molecule Sampling of Dihydrofolate Reductase Shows Kinetic Pauses and an Endosteric Effect Linked to Catalysis. ACS Catalysis (2022). doi/10.1021/acscatal.1c04388
Directional conformer exchange in dihydrofolate reductase revealed by single-molecule nanopore recordings. Nature Chemistry (2020). doi.org/10.1038/s41557-020-0437-0
ChemistryWorld - EurekAlert - NewsBreak - PhysOrg - Bioengineer - eScience News - Medical News - PressReleasPoint - Science Daily - AzoLifeSciences
Engineering and Modeling the Electrophoretic Trapping of a Single Protein Inside a Nanopore. ACS Nano (2019) doi.org/10.1021/acsnano.8b09137
Direct electrical quantification of glucose and asparagine from bodily fluids using nanopores. Nature Commununications (2018) 10.1038/s41467-017-01006-4
-Eurekalert - Sciencelinx - PhysOrg - Science News - Science Daily - Canada News - LongRoom - GEN - R&D - Infosurhoy - Bionity - Health Innovations - Bioanalysis - Kennislink - Chemistry World - LabMedica
Real-time conformational changes and controlled orientation of native proteins inside a protein nanoreactor. JACS (2017) doi: 10.1021/jacs.7b10106. PDF
Single-Molecule Analyte Recognition with ClyA Nanopores Equipped with Internal Protein Adaptors. JACS (2015). DOI
Tuning the Size and Properties of ClyA Nanopores Assisted by Directed Evolution. JACS (2013). http://pubs.acs.org/doi/abs/10.1021/ja4053398
An engineered ClyA nanopore detects folded target proteins by selective external association and pore entry. Nano Letters (2012). http://dx.doi.org/10.1021/nl3024438